1. Text-mining of the literature
To generate a candidate list of neurotrophic/protective proteins using automated text-mining of the literature, current collections of fulltext scientific articles from the PubMed database (Open Access Subset) and the publisher Elsevier were processed using two approaches: (1) A term co-occurrence scoring approach, and (2) a phrase search using regular expressions.
(1) For the initial co-occurrence mining approach, the occurrences of gene/protein named entities and the keywords "neurotrophic" and "neuroprotective" were determined across all collected fulltext articles, and the relative co-occurrences were then scored for each combination of a gene/protein name and the two keywords using the pointwise mutual information (PMI) score, following the approach by Tsuruoka et al. . Specifically, the PMI score is defined as:
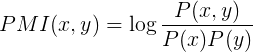
where P(x) is the proportion of the documents that match the query gene/protein name x, p(y) is the proportion of documents that contain the keyword y, and p(x, y) is the proportion of documents that match both the query gene/protein name and contain the keyword. The PMI score reflects how much more often the query gene/protein name and a keyword co-occur in the fulltext articles than expected by chance. For each known human gene covered in the HGNC database and identified in the document collection, a corresponding PMI score was determined.
(2) As a second complementary literature mining approach, a regular expression phrase search was conducted across all collected fulltext articles. For this purpose, the following phrases were searched:
"X is (a) neuroprotective...",
"X has (a) neuroprotective...",
"X is (a) neurotrophic...",
"X has (a) neurotrophic...",
where X must be a gene/protein named entity and text shown in brackets is optional. Fast text searches were implemented using the document search server Apache Solr . In order to combine the results from this phrase search method and the co-occurrence scoring approach into a final ranking of candidate neurotrophic/protective genes, genes were sorted by their PMI scores and by how often they were identified in the phrase searches, and the sum-of-ranks across these two sorted lists determined the final ranking. A preliminary manual inspection of the combined ranking suggested that only genes up to a rank of approx. 150 in the list contained a sufficient proportion of true positive neurotrophic/protective genes to warrant further detailed curation; hence, only the top 150 candidate genes were used for the manual curation, as outlined in section [sec:2].
Powered by Pandoc and www.tsmean.com | Contact: bersling at gmail
2. Manual literature curation
For the manual curation of candidate neurotrophic and neuroprotective genes/proteins obtained from text-mining of the literature, each of the authors received an Excel-file containing a list with a subset of the candidates. These genes were curated according to the following process and guideline:
The goal of the curation was to identify peer-reviewed articles in public literature databases, which either confirm or refute a neurotrophic and/or neuroprotective role for the candidate genes according to unambiguous and non-conflicting experimental evidence reported in the paper, and to annotate the genes with short sentences extracted from the original articles that capture the type of the observed trophic/protective effect and the supporting evidence. As outlined in the main manuscript, the curators were asked to categorize proteins according to the following definitions:
A protein was classified as neurotrophic if experimental evidence in-vitro or in-vivo reported in the peer-reviewed literature confirms that it can specifically augment proliferation, differentiation, growth, and/or regeneration of neurons;
A protein was classified as neuroprotective if experimental or clinical evidence reported in the peer-reviewed literature confirms that it can specifically slow or halt the progression of neuronal atrophy or cell death following the onset of disease or clinical decline in humans or of corresponding established markers in in-vitro/in-vivo disease models.
According to these definitions the curation differentiated between proteins shown to have both neurotrophic and protective functions, only trophic or protective functions, and proteins for which no evidence for trophic/protective effects have been reported so far. The in-vitro and in-vivo evidence for protective and trophic functions was documented separately, and cases in which condition-specific trophic or protective effects have been described were highlighted using a dedicated field in the database. Additional neuroprotective/trophic genes discovered during the curation process were also added to the database if they matched with the above definitions. In order to capture all relevant information for the database entries and filter out false-positives and genes with conflicting or negative evidence for trophic/protective functions, the following detailed step-by-step guideline and procedure was implemented:
Identifying relevant evidence in the literature: To evaluate a candidate gene and identify relevant information in the literature beyond the initial text-mining results, keyword searches in public literature databases (PubMed, ScienceDirect, Scopus and Web of Science) using standard human gene symbols in HGNC format and full gene names were applied for each of the candidates. Gene name aliases derived from the GeneCards database were also taken into consideration in order to resolve possible ambiguities and identify further relevant articles. Combined database queries with AND/OR operators were used to find articles mentioning both neurotrophic/protective functions and discussing relevant in-vitro or in-vivo experimental evidence, according to the following query format: "[Gene name] AND (neurotrophic OR neuroprotective) AND (in-vitro OR cell culture OR in-vivo OR animal model)".
Identifying conflicting/negative evidence: To check whether previously reported protective/trophic functions for a protein have been invalidated by other studies, or conflicting evidence exists, the search queries above were extended to include the keywords/phrases "not neuroprotective", "not neurotrophic", "conflicting", "negative", "false", "invalidated" or "corrigendum". Further false-positive genes were removed by filtering out cytokines wrongly labeled as neurotrophins from the candidate list. Moreover, proteins for which neurotrophic / neuroprotective activities were described to represent an unspecific and non-targeted downstream side effect of modulating the activity of a generic regulatory gene were filtered out (i.e. transcription factors that regulate many diverse genes, including a subset with neurotrophic functions, were be considered as false positive).
Identifying condition-specific trophic or protective activities: To check whether identified neurotrophic/protective functions are reported to be condition-specific, relevant text passages in the identified scientific articles were studied in detail to determine whether a dependency of the function on specific factors, signals or cellular conditions was described. Moreover, additional search keywords were used in the manual curation ("depends on", "controlled / modified / altered / restricted by") to identify condition-specific effects.
Documenting curation results: After inspecting the determined relevant information sources in the literature and collecting the experimental evidence for neurotrophic/-protective functions and/or conflicting evidence, the results were added to the database by providing detailed inputs for the following items/fields in the database:
Gene Symbol (canonical human gene symbol according to the HGNC database )
Full gene name
Activity type: Either "neurotrophic", "neuroprotective", "neurotrophic/neuroprotective" (if there is evidence for both trophic and protective functions), or "no trophic/protective activity identified"
PubMed-IDs or DOIs for articles providing in-vitro evidence for neurotrophic/neuroprotective activities
PubMed-IDs or DOIs for articles providing in-vivo evidence for neurotrophic/neuroprotective activities
PubMed-IDs or DOIs for articles reporting condition-dependent trophic/neuroprotective effects(these may be all or a subset of the identifiers for the previous two items)
Up to two representative sentences from each of the articles that provide in-vitro evidence for neurotrophic/neuroprotective actions
Up to two representative sentences from each of the articles that provide in-vivo evidence for neurotrophic/neuroprotective actions
Up to two representative sentences from each of the articles that report condition-dependent evidence for neurotrophic/neuroprotective actions
